Introduction to Radiomics with Python
What is Radiomics?
Radiomics is a field at the intersection of medical imaging and data science that focuses on extracting large quantities of quantitative features from clinical images. These features can include texture measurements, shape descriptors, intensity statistics, and more. By converting image data into structured information, radiomics aims to reveal patterns that may not be visible to the naked eye, leading to more personalized and data-driven medical decision-making.
Radiomics workflows often involve specialized algorithms and software to systematically analyze medical images—such as CT, MRI, and PET scans—and distill them into meaningful numerical features. These features can then be studied in relation to clinical outcomes, genetic information, or treatment responses, providing insights that can contribute to precision medicine approaches.
Why is Radiomics Important in Radiology?
Traditional image interpretation in radiology relies heavily on subjective or qualitative assessments by expert radiologists. However, as medical imaging technologies have advanced, the volume and complexity of image data have grown dramatically. Radiomics helps address this complexity by providing an objective framework for image analysis, enabling reproducible and high-throughput extraction of features.
This shift from subjective observation to quantitative measurement enhances diagnostic accuracy, aids prognostic evaluations, and offers more detailed treatment monitoring. For instance, subtle textural differences in a tumor—undetectable to the human eye—can be captured by radiomic features. Such features may correlate with tumor aggressiveness, likelihood of metastasis, or even genetic mutations. These capabilities underscore why radiomics is becoming an integral part of modern clinical research and practice.
Brief Overview of How Radiomics Has Evolved
Although quantitative imaging research has been ongoing for decades, the term “radiomics” was popularized in the early 2010s. Early work in quantitative imaging often used a small set of features—like shape or basic intensity measures—to correlate with patient outcomes.
Over time, as computing power and data storage capabilities improved, researchers began to explore larger, more complex feature sets derived from medical images. This led to the development of standardized feature definitions and software packages (e.g., PyRadiomics), which streamlined the radiomics process.
In tandem with advancements in data science and machine learning, radiomics evolved from an experimental research area to a core component of many radiology-focused studies. With the promise of linking imaging features to clinical and molecular data, radiomics started to attract interdisciplinary teams comprising radiologists, data scientists, oncologists, and bioinformaticians. This collaborative environment has accelerated progress and expanded potential applications across various medical domains.
Current Research Directions
Current radiomics research is highly focused on refining feature extraction methods, improving reproducibility, and developing robust predictive models for clinical use. Advanced techniques like foundation models or deep learning-based radiomics enable automated feature extraction that can capture more complex patterns than traditional handcrafted approaches.
Example Radiomics Workflow Overview
The radiomics workflow typically follows a structured series of steps designed to ensure robust and reproducible analyses. The major phases are:
- Image Acquisition: This is the starting point of any radiomics study, where medical images (e.g., CT, MRI, PET) are collected. Proper image acquisition parameters and protocols are crucial for minimizing variability.
- Segmentation: Once images are acquired, a specific region of interest (ROI) must be isolated (e.g., a tumor in oncology studies). Segmentation can be performed manually by expert radiologists or through automated/semi-automated algorithms.
- Feature Extraction: In this phase, radiomic features—such as intensity statistics, textural patterns, and shape descriptors—are calculated from the segmented ROI. Tools like PyRadiomics help automate this process.
- Feature Selection: Because hundreds or even thousands of features can be extracted, it is necessary to select the most relevant ones. Techniques like correlation analysis, recursive feature elimination, or regularization-based methods help reduce dimensionality and prevent overfitting.
- Model Building: Selected features are then fed into machine learning or statistical models to predict clinical outcomes (e.g., survival, treatment response). Approaches can range from logistic regression to more complex algorithms like random forests or support vector machines.
- Validation: Finally, model performance is assessed using standard metrics (e.g., accuracy, ROC AUC, precision/recall) and validation strategies such as cross-validation or external validation cohorts. This step ensures generalizability of the model to new data.
Radiomics Workflow Step-by-Step
Step 1: Region of Interest (ROI) Segmentation
ROI segmentation identifies and isolates a specific region of interest (e.g., a tumor) within a medical image. It ensures that subsequent analyses (such as feature extraction) focus only on relevant tissue, improving the accuracy and clinical value of radiomic analyses.
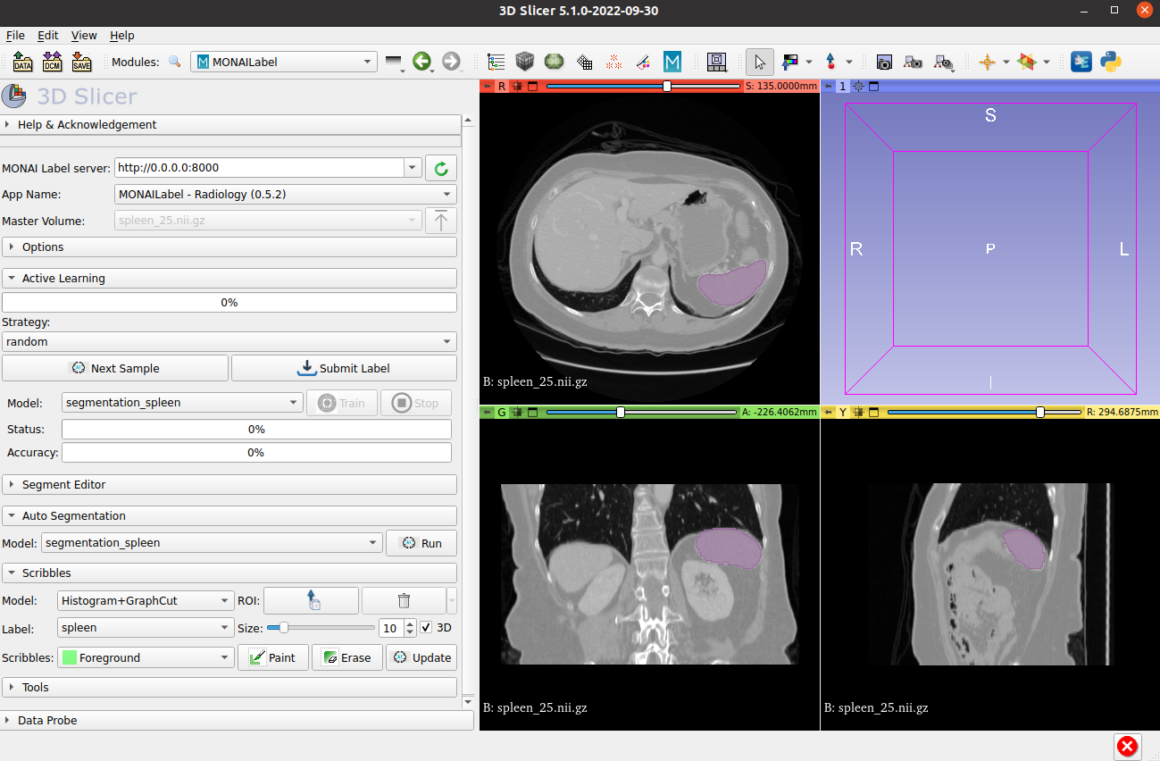
Manual Segmentation
Performed by clinicians or trained radiologists who use software tools to delineate the region of interest (ROI) slice by slice. It offers high accuracy when done by domain experts but can be time-consuming, subjective, and prone to inter-observer variability. In research settings, manual segmentation is often considered a “gold standard” reference when developing or evaluating automated methods.
Automated Segmentation
Uses computational algorithms to identify and segment structures within medical images. Approaches can range from simple thresholding or region growing methods to advanced machine learning or deep learning models. Automated segmentation saves time and reduces operator bias but may require large training datasets for optimal performance. Hybrid or semi-automated methods—where a user provides initial seed points or manual corrections—are also widely used in practice.
Step 2: Feature Extraction with PyRadiomics
Feature extraction involves computing quantitative descriptors (e.g., shape, texture) from segmented ROIs. These features capture patterns that may be invisible to the naked eye, enabling more precise diagnosis, prognosis, or treatment response prediction in medical imaging.
What is PyRadiomics?
PyRadiomics is a widely used open-source Python library designed for high-throughput extraction of radiomic features from medical imaging data. It implements standardized feature definitions, reducing variability across studies and enabling reproducible research. PyRadiomics can handle multiple image and mask file formats (e.g., DICOM, NIfTI) and offers a command-line interface as well as a Python API.
PyRadiomics categorizes its features into several classes, each capturing different aspects of the ROI:
- First-Order Statistics: Basic intensity metrics (mean, median, variance).
- Shape Features: Descriptors of the ROI’s geometry (volume, surface area, sphericity).
- Texture Features: Capturing spatial relationships of voxel intensities (GLCM, GLRLM, GLSZM, etc.).
- Wavelet-Based and Filtered Features: Features extracted from transformed images (e.g., LoG, Wavelet) that highlight specific spatial frequency components.
By combining multiple feature classes, PyRadiomics can generate hundreds to thousands of features from a single ROI, capturing subtle variations that might be clinically relevant. Careful parameter selection is critical for obtaining meaningful and consistent features.
What is SimpleITK?
SimpleITK is a popular library for image processing and analysis, it offers a range of functions for reading/writing medical image formats, filtering, registration, and segmentation. Simple thresholding or region growing can be easily implemented, and the library integrates well with other Python tools such as NumPy and PyRadiomics.
Step 3: Feature Selection
After extracting a potentially large number of features through radiomics, it is crucial to reduce dimensionality and select only the most informative variables. This step helps to enhance model performance, mitigate overfitting, and improve interpretability. Feature selection techniques generally fall into three categories: filter methods, wrapper methods, and embedded methods.
Filter methods evaluate features based on their intrinsic statistical properties or relationships with the target variable, without involving a specific predictive model. Advantages of filter methods include simplicity and speed, as they do not require training a predictive model. However, they may overlook nonlinear or context-specific relationships that a model-based approach could capture.
Wrapper methods can yield high-performing feature subsets because they directly optimize for the predictive task. However, they can be computationally expensive, especially when dealing with very large feature sets, as each iteration involves model training.
Embedded methods integrate feature selection into the predictive model’s training process. They can include tree-based models (e.g., random forest, gradient boosting), which inherently rank features by their importance during training. Embedded methods offer a good balance between performance and computational cost, as they select features while simultaneously learning model parameters.
By carefully applying feature selection techniques, radiomics researchers can reduce model complexity, highlight the most clinically relevant features and build more generalizable predictive models.
Step 4: Model Building
Once you have extracted a robust set of radiomic features, the next essential step involves selecting and training a predictive model. The choice of algorithm depends on factors such as dataset size, feature dimensionality, and the nature of your prediction task (e.g., classification vs. regression).
Many researchers find a Random Forest classifier a good starting point because it can handle large numbers of features, offers built-in measures of feature importance, and is relatively resilient to overfitting when appropriately tuned. However, exploring different machine learning methods—like support vector machines or gradient boosting approaches—is crucial to finding the best fit for your data.
Regardless of the algorithm chosen, the model-building phase involves feeding your radiomic features (often combined with clinical or demographic data) into the machine learning algorithm and adjusting parameters or hyperparameters to maximize performance. By experimenting with parameter settings, you can systematically identify which configurations yield the strongest predictive power.
Keeping track of these configurations and maintaining consistent data preparation steps is essential. In doing so, you can ensure that any improvements in model performance are genuinely due to better algorithmic settings and not inadvertent changes in how data were handled.
Step 5: Validation
After training, it’s vital to evaluate how well the model generalizes to data it has never encountered. This is typically achieved by splitting your dataset into separate training and test subsets or by using more advanced methods like cross-validation.
Suppose a model performs exceptionally on the training data but fails to maintain that accuracy on unseen data. In that case, it may be overfitted—meaning it has “memorized” training examples rather than learning generalizable patterns.
Validation goes beyond simply calculating overall accuracy or error rates. Detailed evaluations often include metrics such as precision, recall, F1-score, or area under the receiver operating characteristic curve (AUC), depending on whether your problem is classification or another outcome prediction.
A confusion matrix can further reveal if the model systematically confuses one category with another. By carefully analyzing these results, you can refine your feature set, adjust hyperparameters, or even switch algorithms to improve reliability. Ultimately, robust validation ensures that your radiomics model not only fits the data you have but also holds meaningful predictive value in real-world clinical or research scenarios.
Next Step
In the next post, we will develop a comprehensive radiomics application using Python to classify breast ultrasound lesions, a critical task in breast cancer detection and diagnosis. Utilizing Python’s robust ecosystem of libraries—such as PyRadiomics for feature extraction, SimpleITK for image processing, and scikit-learn for machine learning—we will systematically process ultrasound images to extract meaningful features from the regions of interest (ROIs). These features will then be used to train and validate machine learning models that can accurately classify lesions, potentially enhancing diagnostic accuracy and aiding radiologists in making informed decisions.
Throughout this application development, we will learn essentials in data preprocessing, feature selection, model training, and validation to ensure the reliability and clinical relevance of our classification system. By the end of this project, you will have hands-on experience in building a radiomics-based diagnostic tool, demonstrating the powerful synergy between medical imaging and data science in advancing personalized medicine.